
Targeted gene expression in cell-specific patterns
The GAL4 system is one of the most powerful tools for targeted gene expression. It allows for temporal and spatial control of gene expression in vivo and has been adapted for many experimental uses. Click for more information and reagents.

Mapping long noncoding RNA-DNA interactions in vivo
RNA DamID combines Targeted DamID with the MCP-MS2 system to profile long noncoding RNA (lncRNA) binding sites in the genome in specific cell types. Click for more information and reagents.

Mapping protein-DNA interactions in vivo
Targeted DamID (or “TaDa”) profiles genome-wide protein-DNA binding in vivo in a cell-type-specific way without cell purification. DNA adenine methyltransferase (Dam)-fusion proteins are expressed at very low levels with both spatial and temporal control. Click for more information and reagents.

Mammalian Targeted DamID
We have modified TaDa to generate mammalian TaDa (MaTaDa), which can be used to identify genome-wide protein/DNA interactions in 100 to 1000 times fewer cells than ChIP-based approaches. Click for more information and reagents.
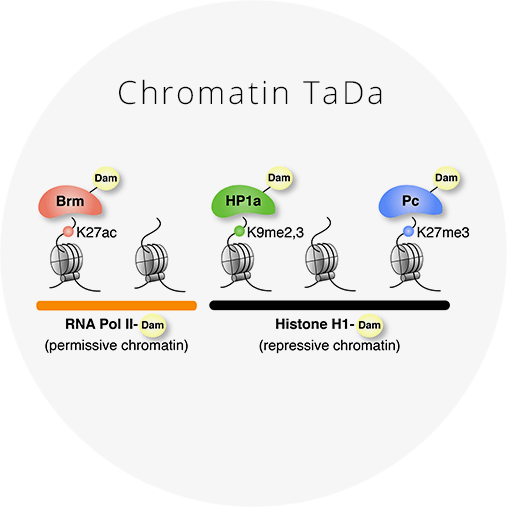
Determining chromatin states in specific cell types in vivo
Chromatin TaDa uses Targeted DamID to assess the genome-wide occupancy of chromatin-binding proteins in vivo. Chromatin states can be determined by profiling RNA Pol II, Brm, HP1a, Pc and Histone H1 binding. Click for more information and reagents.

Tools for preparing Targeted DamID samples and data analysis
Click for the latest molecular biology protocols and analysis pipelines for Targeted DamID.

